April 4, 2016

Selecting genetics to help the U.S. swine herd fight infectious diseases would provide a major financial boost to hog producers everywhere.
Recently, the use of genetics to select or develop pigs that are less susceptible or resistant to the porcine reproductive and respiratory syndrome virus has been extensively reported on in the National Hog Farmer. The 2015 Research Review in the December issue included a report, “Genetic marker may show PRRSV promise,” on the identification by our group and collaborators at Kansas State University (Bob Rowland), USDA-Agricultural Research Service (Joan Lunney), and the University of Alberta (Graham Plastow) of a major gene on pig chromosome 4 for susceptibility to PRRS virus infection through the PRRS Host Genetics Consortium.
Although pigs that have the favorable genotype for this major gene or its linked genetic marker (i.e., pigs that have the AB or BB genotype rather than the AA genotype; the B allele appears dominant, so only one copy of the B allele is needed to obtain the favorable effect) do get infected when exposed to the virus, have lower levels of viremia following infection and maintain a higher growth rate (see Figure 1).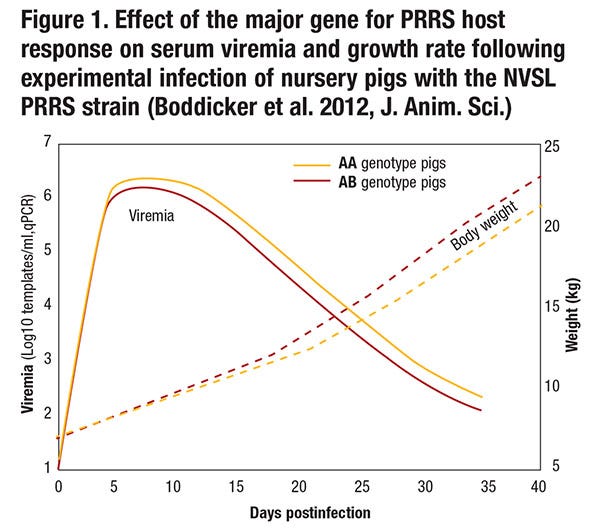
The effects of this gene are the result of a naturally occurring mutation of the GBP5 gene, which produces a protein that is involved in innate immune response. Because of this mutation, pigs that have the unfavorable genotype (AA) produce a truncated GBP5 protein, which hampers their innate immune response to PRRS infection.
Innate immunity refers to the first line of defense to pathogens, in contrast to adaptive immunity, which involves the production of antibodies that are specific to a pathogen. Adaptive immunity results in the “memory” that is developed as a result of vaccination.
The January issue of National Hog Farmer included reports on the successful development of a pig that appears completely resistant to the PRRS virus by the lab of Randall Prather at the University of Missouri. This pig was developed by “editing” the CD163 gene, which produces a protein that the virus uses to enter the pig’s macrophage cells. By blocking this entry, the virus is not able to infect a pig and replicate, resulting in complete resistance.
This is a promising development, but requires substantial additional research to ensure its efficacy and absence of negative side-effects. In addition, pigs produced by gene editing have not yet been approved for use in pork production.
Complete resistance to a pathogen is an exception, rather than the rule. Even for the PRRS-resistant pig, the question remains whether it is resistant to all variants of the PRRS virus. In addition, pathogens evolve rapidly and could mutate or recombine into strains that evade whatever resistance mechanism the pig employs.
Also, it will likely be impossible to develop pigs that are completely resistant to all major diseases. Thus, continued emphasis on developing strategies to deal with cases of incomplete resistance is needed, which is the focus of this article. In addition, although vaccination can be an important tool to combat viral disease, vaccines are often not completely effective. Thus genetic approaches to combat disease are needed to complement vaccination, biosecurity and eradication approaches.
Coping with infection
There are several mechanisms that an animal can use to prevent infection or to cope with infection.
Resistance refers to the ability of an animal to prevent infection when exposed to a pathogen or to limit replication of the pathogen when infected. The gene-edited pig described above is an example where resistance appears to be complete, but in most cases resistance is by degree and can be quantified by measuring pathogen load in animals that have been exposed to the pathogen. Thus, pigs with the favorable genotype for the major gene on chromosome 4 that we identified are more resistant than pigs with the unfavorable genotype because they have lower levels of viremia following infection (see Figure 1). This could be because more resistant animals are better able to block entry of the virus, to reduce replication of the virus, to prevent release of the virus, or to kill the virus. Although the difference in viremia between AA and AB pigs appears small, it is highly significant and also has biological significance because pigs with the favorable genotype grow faster.
Tolerance refers to the ability of an animal to maintain performance at a given level of infection or pathogen load. Thus, at a given level of viremia, growth and performance of tolerant animals are less affected than that of less-tolerant animals. This could be because tolerant animals are better able to deal with the damage that is done by the pathogen or because they are more effective at mounting an immune response, diverting less energy away from performance.
Whereas tolerance refers to the impact on performance of a given pathogen load that is present in the animal, the concepts of resilience and robustness refer to the ability of an animal to maintain performance upon exposure to a pathogen. Thus, resilience combines resistance and tolerance.
To further illustrate the concepts of resistance, tolerance and resilience, consider the relationship between level of exposure to a pathogen and pathogen load for the progeny of two boars, A and B, that is shown in Figure 2a. Here pathogen load could represent the level of viremia measured in blood following exposure. The graphs show that, for a given level of exposure, progeny of boar B have a pathogen load that is twice as large compared to progeny of boar A. This reflects differences in resistance to the pathogen — progeny of boar A are more resistant.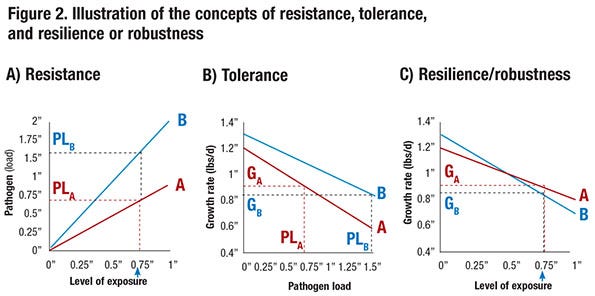
Figure 2b shows the relationship between pathogen load and growth rate. At a pathogen load of zero — i.e., in a clean environment — progeny of boar B have a 0.1 pound greater growth rate per day than progeny of boar A. In addition, as pathogen load increases, growth rate drops more quickly for progeny of boar A than for progeny of boar B; at a pathogen load of 1 (in arbitrary units), the growth rate of progeny from boar A has dropped 0.4 pound per day (from 1.2 to 0.8), compared to a drop of only 0.3 pound per day for progeny of boar B (from 1.3 to 1.0). This reflects tolerance — i.e., for a given level of pathogen load (e.g. viremia), the growth rate of the progeny of boar B is less affected. In other words, the progeny of boar B are more tolerant. Thus, if the progeny of these two boars have the same pathogen load, boar B would be preferred. The pathogen load that an animal reaches upon exposure, however, depends on its resistance.
For example, consider an exposure level equal to 0.75. Based on Figure 2a, this results in a pathogen level equal to 1.5 for boar B (PLB) but of only 0.75 for the more resistant boar A (PLA). At these respective pathogen levels, progeny of boar A are expected to grow 0.9 pound per day (GA = 0.9 at PLA = 0.75). However, because progeny of boar B are less resistant and have twice as high a pathogen load (PLB = 1.5) at this level of exposure, they are expected to grow only GB = 0.85 pound per day. Thus, although progeny of boar B have a higher growth rate without exposure and are more tolerant to the pathogen, they have a lower growth rate at this exposure level because they are less resistant.
Combining Figures 2a and 2b, Figure 2c shows the relationship between level of exposure and growth relate, i.e., resilience. Thus, at a level of exposure greater than 0.5, progeny of boar A have greater growth rate than progeny of boar B, although progeny of boar B have a greater growth rate without exposure. The level of performance at a given level of exposure is referred to as resilience or robustness. Thus, at a level of exposure greater than 0.5, boar A is more resilient or robust than boar B.
Assuming that development of pigs that have complete resistance to all major diseases is not achievable, the ultimate goal is to develop pigs that are resilient or robust given the level of exposure that is expected in the field. Based on the concepts introduced above, this can be achieved in two ways — by improving resistance or by improving tolerance.
If disease eradication is the goal, then emphasis should be on improving resistance, because improving tolerance does not change the extent to which a pathogen can infect and replicate in the host.
However, increasing resistance puts more selective pressure on the pathogen to evolve, which may be counterproductive in the long term. Resistance, however, also reduces shedding and, therefore, has an added advantage by reducing infection pressure in the population.
The best strategy for improvement likely differs between pathogens but may be to focus on improving a combination of resistance and tolerance.
Generalized vs. specific
Another important concept is that of generalized versus specific immunity. “Generalized immunity” refers to the ability to respond to a range of disease challenges. In contrast, “specific immunity” refers to the ability to respond to a specific disease challenge.
Vaccines by definition focus on developing an animal’s specific immunity. This also means that a different vaccine is needed for each pathogen and perhaps even for different strains of the same pathogen (e.g., for heterologous PRRS virus strains). In addition, effectiveness of a vaccine may change as the pathogen evolves.
Because genetic improvement is a long-term process, the goal of genetic improvement should be to improve generalized immunity, rather than specific immunity. This often means that genetic improvement should focus on components of the immune system that impact classes of diseases, rather than a specific disease. Components of the innate immune system would be prime candidates. This could also include the ability of an animal to mount an effective adaptive immune response. Targeting generalized immunity reduces the chance that the pathogen will evolve around it and also reduces the possibility that improvement of response to one disease will be unfavorable for other diseases.
Effect of a major gene
The major gene for host response to PRRS on chromosome 4 that was described above is an important finding, in particular, because it appears that having at least one copy of the favorable B allele provides some level of protection against both heterologous strains of the PRRS virus that we have tested through experimental infections so far. These results are, however, based on a challenge model in clean facilities, using naïve pigs and a specific PRRSV strain. Disease in the field is obviously much less controlled and often occurs in pigs with prior or concurrent exposure to other pathogens.
To address these concerns, we have embarked on several studies in collaboration with the groups of Rowland at KSU and Lunney of USDA in Beltsville, Md., with funds from USDA-National Institute of Food and Agriculture and in close collaboration with Choice Genetics and PIC. These include studies in which groups of 200 nursery pigs are co-infected with field isolates of PRRS virus and PCV2b at KSU, as well as field studies in which groups of 200 clean nursery pigs are introduced into barns that have a history of health challenges or that are primed with seeder pigs from barns with health problems.
To date, the co-infection studies have shown that although effective vaccines for PCV2b are available, co-infection with PRRS and PCV2b is a good model for combinations of diseases that occur in the field. In addition, in these co-infection trials half of the piglets are vaccinated with a modified-live PRRS vaccine 28 days prior to co-infection to evaluate the impact of vaccination and to determine whether we can identify pigs that respond better to these vaccines. Results showed that, compared to AA pigs, AB pigs had significantly lower PRRS viremia levels, both following vaccination and following co-infection, confirming the positive effect of the B allele on resistance.
Numerically, the effect of the major gene was greater upon first exposure to PRRS, which is consistent with the role of the GBP5 gene in innate immunity. AB pigs also grew faster following vaccination but we found no difference in growth rate between AA and AB pigs following co-infection, with or without vaccination.
Interestingly, the major gene for PRRS had no effect on PCV2b viremia levels in pigs that were not vaccinated, but AB pigs did have a significantly lower level of PCV2b viremia in pigs that were first vaccinated for PRRS. This demonstrates the immunological interactions that take place when animals are exposed to multiple pathogens and the protective effect of the major gene for PRRS in these multi-pathogen scenarios. Taken together, these results show that selecting pigs that have the favorable genotype for this major gene (AB or BB) is a promising strategy to select for improved response to not just PRRS infection, but also co-infection with PCV2b and, perhaps, other pathogens. The ongoing field studies are generating data to confirm these findings in commercial barns.
Challenges to improvement
It is well-known that traits related to resistance to disease and how a pig responds and performs in the face of a disease challenge have a sizable genetic component, and we have clearly demonstrated that in the PRRS host genetics consortium studies. A major challenge to selecting pigs that are more resistant or less susceptible to disease is how to identify such pigs.
A logical solution would be to collect data on animals that are exposed to disease. This is, however, not possible in most breeding programs because the nucleus breeding herds of those programs must be maintained under the highest biosecurity provisions. As a result, most genetic improvement is based on data from pigs that are kept in high-health conditions. Referring back to Figure 2c, this would mean that we would be selecting boar B because it has the best performance in a clean environment (with zero exposure). However, if most of the pigs that descend from this boar are in a commercial environment with a substantial pathogen load, then boar A would be the preferred boar because his progeny perform better in such an environment.
Thus, an important question for breeding organizations is how to obtain information that can be used for selection of their nucleus animals for disease resistance, tolerance or resilience. Possibilities include:
■ Collect performance data at the commercial level on progeny or sibs of nucleus animals. Although several breeding organizations have embarked on this, collecting data at the commercial level that can be tied back to nucleus animals is a major challenge because pedigree information is often lacking.
■ Use genomic prediction based on commercial performance. Estimates of marker effects could be obtained by training on records of performance obtained on commercial animals in the field (see Figure 3). Although this does in principle not require pedigree information on the commercial animals, this option would involve substantial genotyping costs.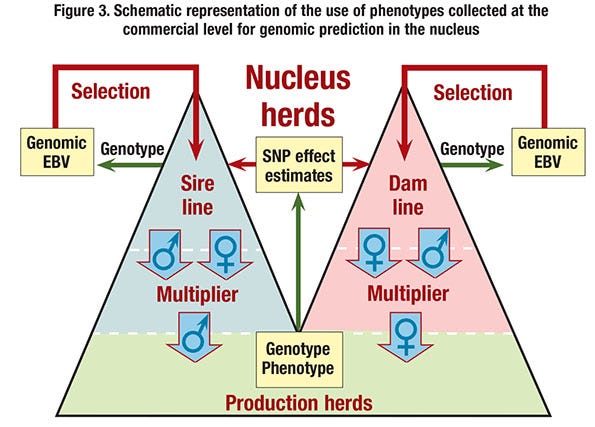
■ Select genetic markers with major effects on host response to disease resistance. The major gene for PRRS described previously is a good example how a major gene can be used to then select nucleus animals based on their genotype at this gene or a nearby marker. Most disease traits are, however, affected by many genes with small effects, making the prospects for this option limited.
■ Develop indicator traits of immune response that can be measured on nucleus animals. The idea here would be to develop immune response tests that can be applied to animals in the nucleus without jeopardizing biosecurity. Such tests have, for example, been developed for dairy bulls by Bonnie Mallard at the University of Guelph in Canada, and are now used by Semex Canada to test, select, and market so-called Immunity+ bulls. Similar tests are currently under development for pigs.
While advances are being made in all these areas, the remainder of this article describes two studies using field data that have the potential to result in indicator traits that can be used to select nucleus animals for improved resistance, tolerance or resilience.
PRRS outbreak herd study
In a recently published study (Serao et al., J. Animal Sci. 2014), we found that PRRS-specific antibody levels in sows following a PRRS outbreak can be used to identify genetics that are more resilient to reproductive PRRS. This study used data from a natural PRRS outbreak in a purebred Landrace multiplier herd from an industry collaborator in Canada.
Following the outbreak, the herd was closed but continued operation, and a veterinary team was organized to collect blood samples approximately 45 days after the start of the outbreak on approximately 640 sows that were in the herd at the time of the outbreak. These samples were sent to a diagnostic lab to quantify the level of PRRS-specific antibodies using a commercial ELISA test. Antibody levels were found to be quite heritable, meaning that daughters of the same boar tended to have similar PRRS antibody levels.
More specifically, we found that ~45% of the differences in antibody levels could be attributed to genetics, which is higher than the heritability we typically find for growth rate in finishing pigs. Moreover, genes in the major histocompatibility complex, which are associated with antibody production, were found to explain a substantial proportion of these genetic differences.
As indicated, this herd continued operation, and all reproductive performance records were collected on the sows that were in gestation at the time of the outbreak. These data were then used to estimate genetic correlations between antibody level and reproductive performance, and we found them to be quite high (~0.7) and favorable, although standard errors of these estimates were substantial because of the still limited amount of data. These high favorable genetic correlations do suggest, however, that daughters of boars that have high antibody levels following exposure to the PRRS virus are more resilient, in that they tend to have fewer stillborn piglets and mummies during the outbreak. This is a promising result, but because of high standard errors, it requires further validation, which we are in the process of obtaining.
Gilt acclimation study
In a related large-scale study, also funded by Genome Canada, the Canadian Swine Health Board and PigGen Canada, and in collaboration with members of the PigGen Canada consortium, over 1,600 clean gilt replacements from six genetic sources and 17 multipliers were introduced into 22 commercial farms across Canada with a history of health challenges. Gilts were entered into the standard acclimation protocol that was in effect in each of these herds, which varied considerable between herds and included vaccination and exposure to seeder or sentinel pigs. Blood samples were taken on all gilts at multiple times during and after acclimation.
Despite the different acclimation protocols and the different environments that these gilts were introduced into, we found that PRRS antibody levels were reasonably heritable (~30%) in blood samples that were collected ~40 days after entry into the acclimation protocol. In addition, estimates of genetic marker effects on antibody levels that were obtained from these data were found to be highly predictive of the antibody levels on sows in the PRRS outbreak herd that was described earlier.
This further confirms the genetic basis of antibody responses to PRRSV in commercial herds and the possibility that antibody levels evaluated following vaccination against PRRS could be used as genetic indicators for reproductive performance during a PRRS outbreak. Reproductive data on these gilts is currently being collected and will be used to estimate relationships between antibody levels during acclimation and subsequent reproductive performance.
Conclusions
How pigs cope with infectious disease has a substantial genetic component and can, therefore, be improved by genetic selection if pigs that have the right genetics can be identified. Therein, however, lies the problem.
First of all, there are several mechanisms that can result in pigs responding better in the face of disease, including resistance and tolerance. Although complete resistance may be the ultimate goal, it will likely be impossible to develop pigs that are completely resistant to the main infectious diseases that pigs are exposed to. In addition, pathogens can evolve much more quickly than we may be able to change the pig. Tolerance, resilience and robustness refer to the ability of a pig to maintain performance in the face of disease and may be a more attractive target for genetic improvement.
The second challenge is how to obtain information that can be used to identify pigs that cope better with disease. Promising strategies in that regard are genetic markers or genomic prediction, as are indicator traits, such as immune response to vaccines or other inert compounds. This is an active area of research, which is expected to result in some effective strategies in the coming years.
You May Also Like



